dataset
Type of resources
Keywords
Contact for the resource
Provided by
-
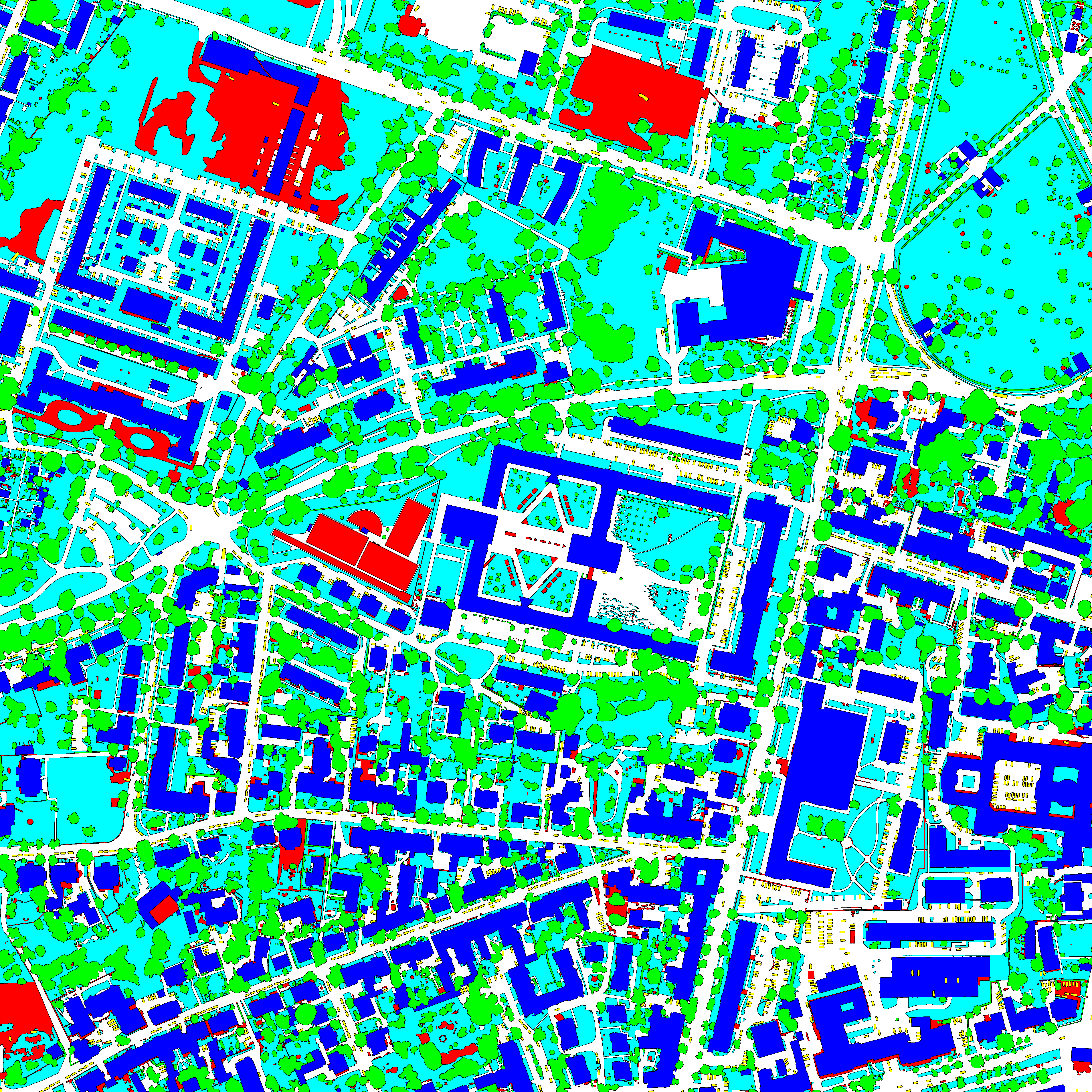
This dataset contains semantic segmentation train-validation-test data splits of (1) the ISPRS Potsdam orthomosaic (https://www.isprs.org/education/benchmarks/UrbanSemLab/2d-sem-label-potsdam.aspx), (2) the RIT-18 landcover orthomosaic (https://github.com/rmkemker/RIT-18), and (3) the industrial environment of the photorealistic Flightmare quadrotor simulator (https://github.com/uzh-rpg/flightmare). All splits are generated by simulating UAV missions at fixed altitudes. We use these datasets in our "An Informative Path Planning Framework for Active Learning in UAV-based Semantic Mapping" paper. Further, it contains (4) model checkpoints of our proposed Bayesian ERFNet framework (https://github.com/dmar-bonn/bayesian_erfnet) pre-trained on Cityscapes, (5) the ISPRS Potsdam and RIT-18 RGB and semantically labelled orthomosaics, and (6) the Flightmare render binary for the industrial environment.
-
Reflectance orthomosaics of PhenoRob Central Experiment derived from 2021 multispectral image data.
-
The dataset RGB-MiniplotBarley contains 22164 RGB images of 11 time points 21. jun - 7. jul 2022 of two genotypes of spring barley grown under single-nutrient deficiencies of N, P, K, S, B and one multi-nutrient deficient treatment from miniplot fertilizer trial at Campus Klein-Altendorf of University of Bonn. The images are annotated with plot number, genotype and fertilizer management and have been taken for a Deep Learning approach for nutrient deficiency recognition.
-

A sweet pepper dataset which was captured in the CKA glasshouse using PathoBot. It contains differently coloured sweet peppers in various ripening stages. More information, citations and a related previous dataset can be found at http://agrobotics.uni-bonn.de/sweet_pepper_dataset/
-
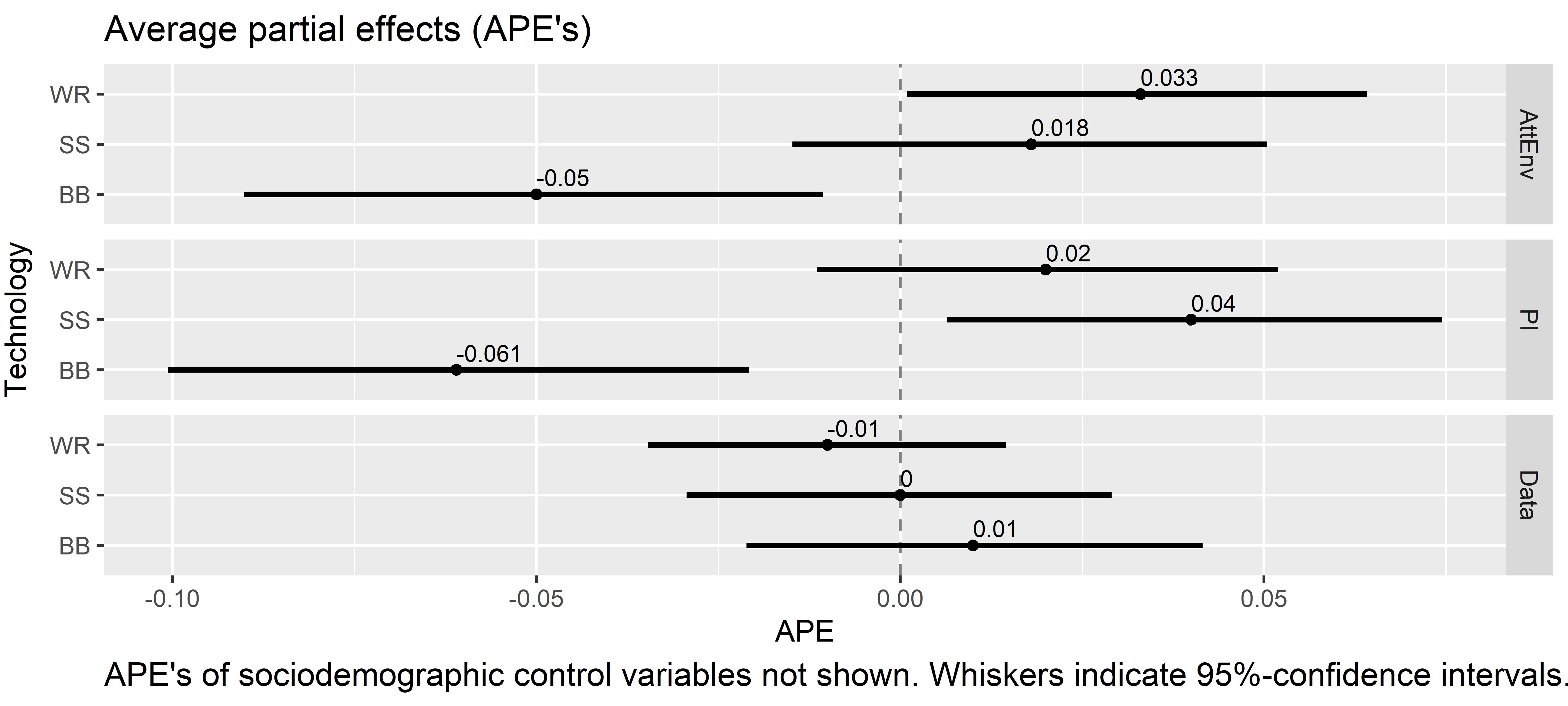
This data set contains survey data from a framed field experiment that was conducted with German crop farmers between February and April 2022. Major components of the data set: a set of psychographic/attitudinal items, results from an economic business simulation game, sociodemographic and farm structural variables (n=334 after data cleaning).
-

Shape completion dataset for sweet peppers. A detailed description of the dataset can be found in our technical report: https://www.ipb.uni-bonn.de/wp-content/papercite-data/pdf/magistri2024arxiv.pdf We additionally provide a small set of python script to load this data: https://github.com/PRBonn/shape_completion_toolkit
-

Data represent an intercropping field experiment carried out at CKA in the year 2021. Shoot and root data collection was conducted with one faba bean cultivar and two spring wheat cultivars sown at three sowing densities. FTIR spectroscopy was used to define the root masses of the two species.
-

This data set contains online survey data from an experiment investigating German public attitudes towards agricultural robots. Major components are the data set containing 2,269 complete questionnaires (after data cleaning), the according Stata do-file to analyze the data and the output files from the preceding construct validity tests in SPSS.
-
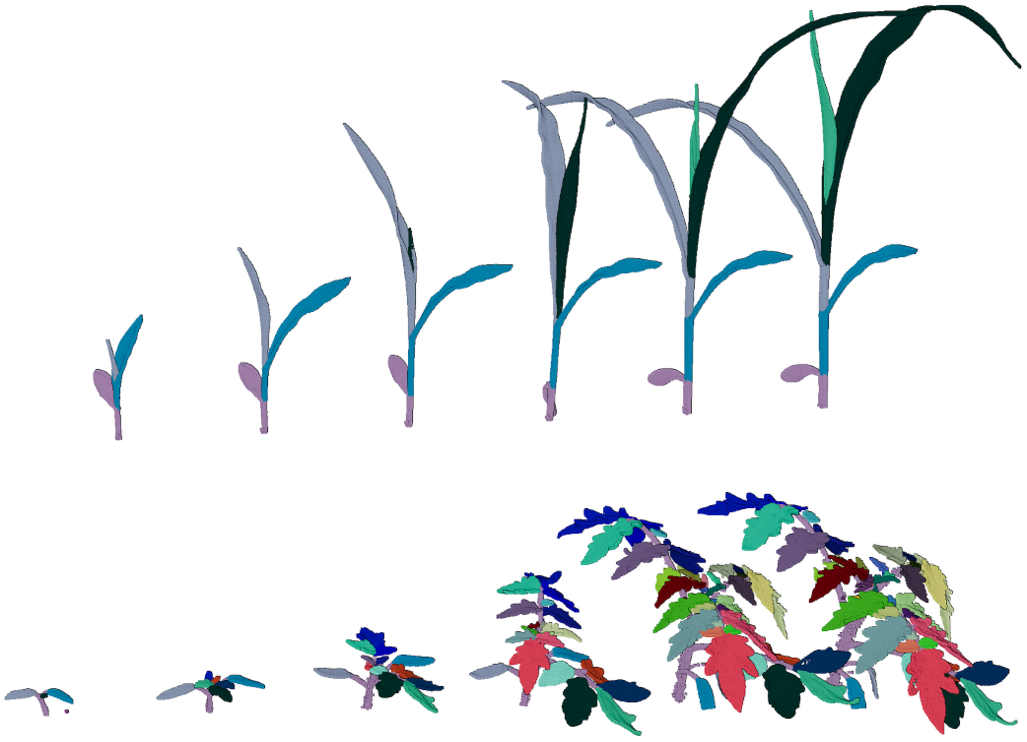
The dataset contains 7 maize plants measured on 12 days. This gives 84 maize point clouds (about 90 Mio. points). From these, 49 point clouds (about 60 Mio. points) are labeled. Furthermore, the dataset contains 7 tomato plants measured on 20 days (about 350 Mio. points). This gives 140 point clouds from which 77 point clouds (200 Mio. points) are labeled. Note that we provide temporally consistent labels for each point in the clouds. We provide labeled and unlabeled point clouds, the file name indicates whether the point cloud is annotated or not. For example, M01_0313_a.xyz is labeled, M01_0314.xyz is not labeled. For the tomato plant point clouds, each annotated file contains the x,y,z coordinates, and the label associated with the point. For the maize point clouds. Each file annotated contains the x,y,z coordinates, and the 2 labels associated with the point. For both species, if no labels are provided, the files contain only the x,y,z coordinates. Cite: D. Schunck, F. Magistri, R. A. Rosu, A. Cornelißen, N. Chebrolu, S. Paulus, J. Léon, S. Behnke, C. Stachniss, H. Kuhlmann, and L. Klingbeil, “Pheno4D: A spatio-temporal dataset of maize and tomato plant point clouds for phenotyping and advanced plant analysis,” PLOS ONE, vol. 16, iss. 8, pp. 1-18, 2021. doi:10.1371/journal.pone.0256340.
-

This dataset contains point clouds of sugar beet plants in field conditions. The data was recorded at Bundessortenamt and was manually labeled for leaf instance segmentation. If you want to use the dataset please contact me.
 PhenoRoam
PhenoRoam